Request for samples
The request must be approved by the St. Jude Comprehensive Cancer Center Hematological Malignancies Program pre-clinical committee. Once the request is approved an MTA will be established between St. Jude (and, where applicable, the institution or cooperative group that provided the primary material) and the requesting institution. The shipping address will be confirmed and one frozen vial per requested sample will be shipped on dry ice along with the protocol to propagate xenografts (using the requester’s shipping account number).
We will endeavor to respond to each request within a week. The exact timeline to receive samples will depend on the following:
- Number of vials we have of the requested sample. If a sample requires propagation on our end, the timeline will typically be several weeks, depending on the growth kinetics of the tumor. Time to engraftment of the primary xenograft is listed on the PROPEL sample search page. However, more than 85% of samples are available for immediate release without further propagation and expansion at St Jude.
- Length of time required to establish an MTA with the requesting institution.
Our aim is to share samples to propel research in the field. To reduce the risk of depleting valuable samples and to avoid issues involved in shipping large number of cryovials, we ask that requests are limited to 5 samples. Scientifically justified requests for more than 5 vials will require consultation and approval. Please detail reasons for larger requests in the comments section of the request form.
One frozen vial each of the requested PDXs will be shipped on dry ice along with instructions for optimal thawing and propagation conditions. Each vial will typically have 20-40 x 106 cells. The requester can propagate and expand the cells in his or her lab.
Primary and xenografted cells typically show poor proliferation in vitro, particularly after thawing. It is recommended that the requester propagate and expand cells in vivo for their own stocks. When shipped, samples will come with instructions for optimal thawing and propagation conditions.
Include the question in the request form or email directly at propel@stjude.org. If you are not able to successfully propagate the cells despite following the protocol we share, we will help you troubleshoot. This may or may not involve shipping another vial of the requested xenograft. After successful propagation, validation of xenografts will be the responsibility of the requester.
PDX material can only be transferred with an MTA signed between St. Jude (and where applicable, the institution or cooperative group that provided the primary material) and the requesting institution. This helps clarify the ownership conditions and specifies how the material can be used.
Please describe PROPEL as an initiative of St. Jude Children’s Research Hospital and cite the URL https://propel.stjude.cloud.
No. All materials received from PROPEL must remain with the requesting investigator, and may not be shared with other investigators at the same institution or elsewhere without explicit approval. Please encourage other interested researchers to contact us directly by filling out the request form.
Explore genomic data
The majority of primary diagnosis and corresponding xenograft samples have been subjected to verification of key oncogenic lesions (e.g. rearrangements and fusions) and/or transcriptome and whole genome sequencing. Genomic data may be interrogated at the PROPEL genomic webpage linked to the PROPEL sample search page. Sequencing data are continually added to this resource. In general, the primary diagnosis and 1-2 representative xenografts are sequenced. Verification of the presence of specific genomic lesions in any sample are the responsibility of the requestor.
- Navigate to the Sample grid under Explore Data
- Search/filter for a sample
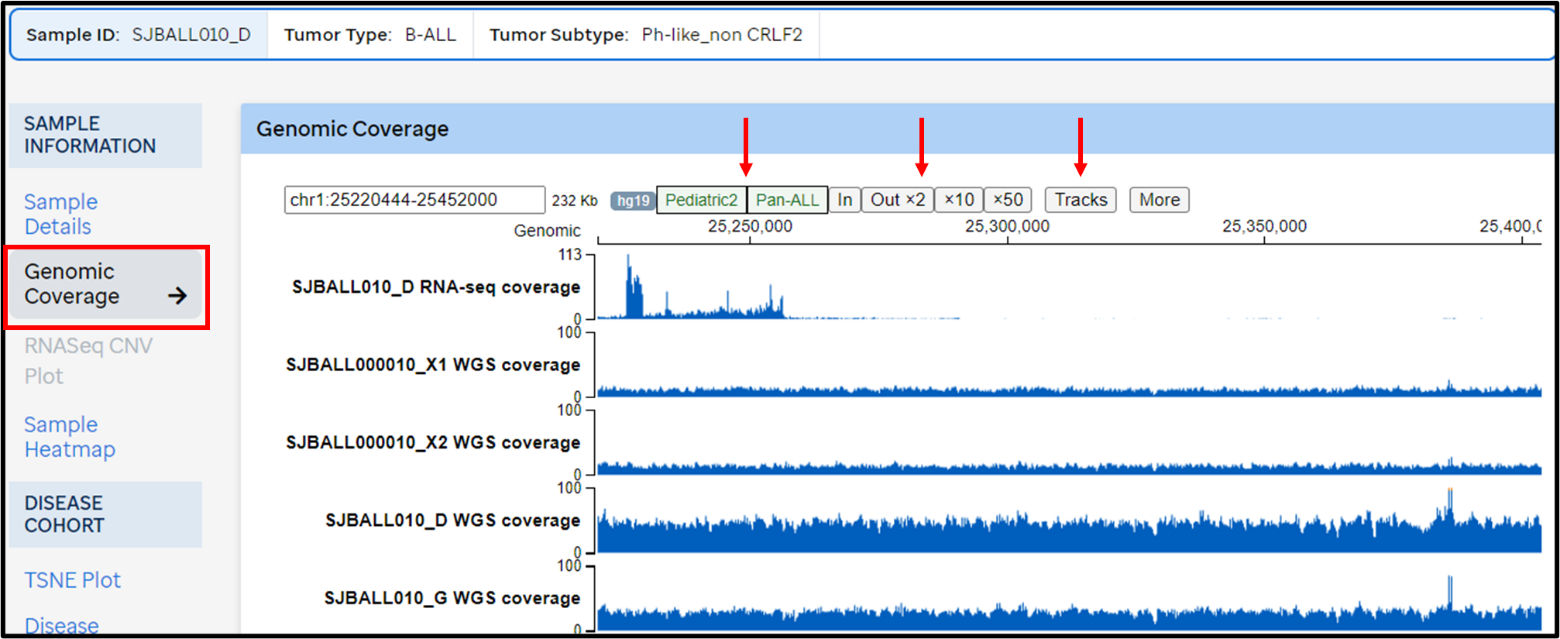
- Click on the sample ID to reveal detailed information (including genomic coverage)
- Genomic coverage provides raw coverage of WGS, WES and RNAseq data for that sample
- The genomic regions can be zoomed in/out, similarly to UCSC browser
- Pediatric mutation data from Pediatric Cancer Genome Project (PCGP) phase II as well as B-ALL mutation and expression data in Pan-ALL study can be viewed
- Users can also add or remove tracks using Tracks tab
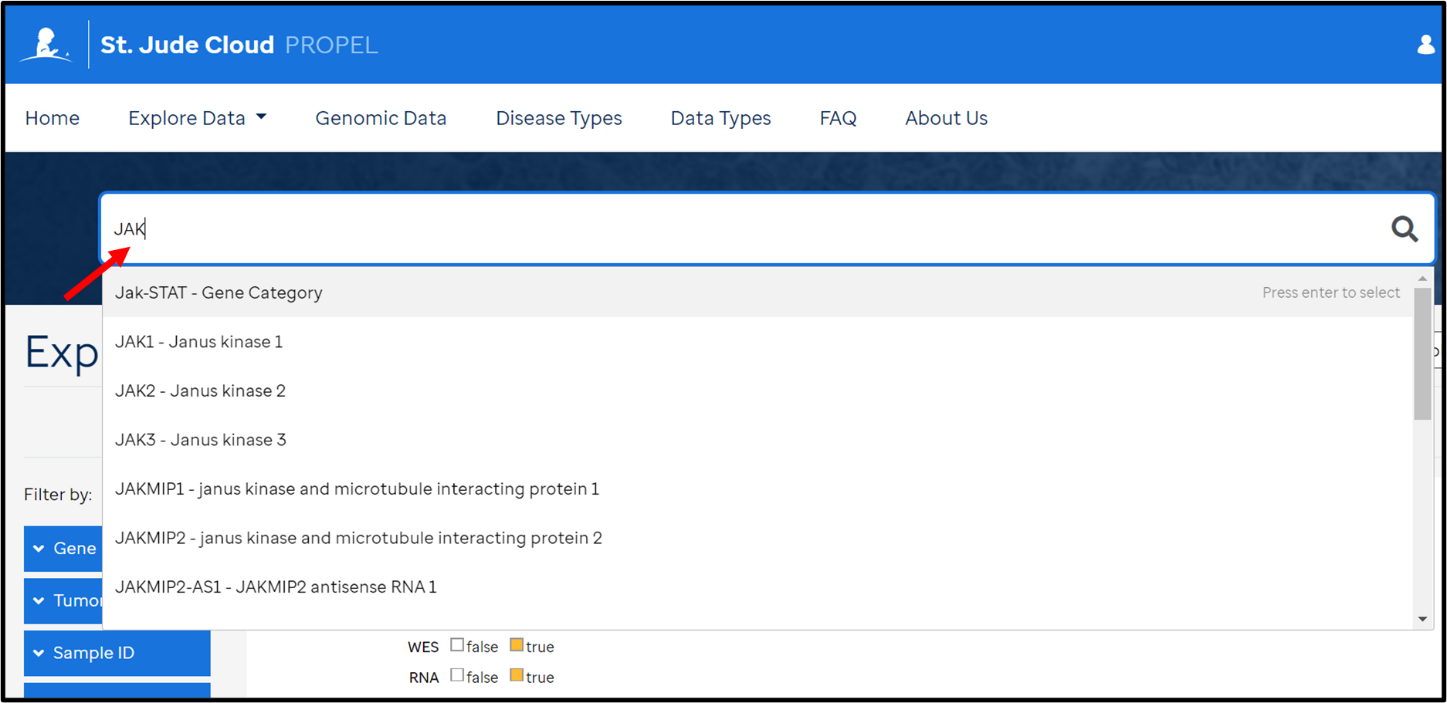
- Navigate to the Genomic Data landing page
- Search for gene(s) of interest using the search box on top OR by using the filter options in the left panel
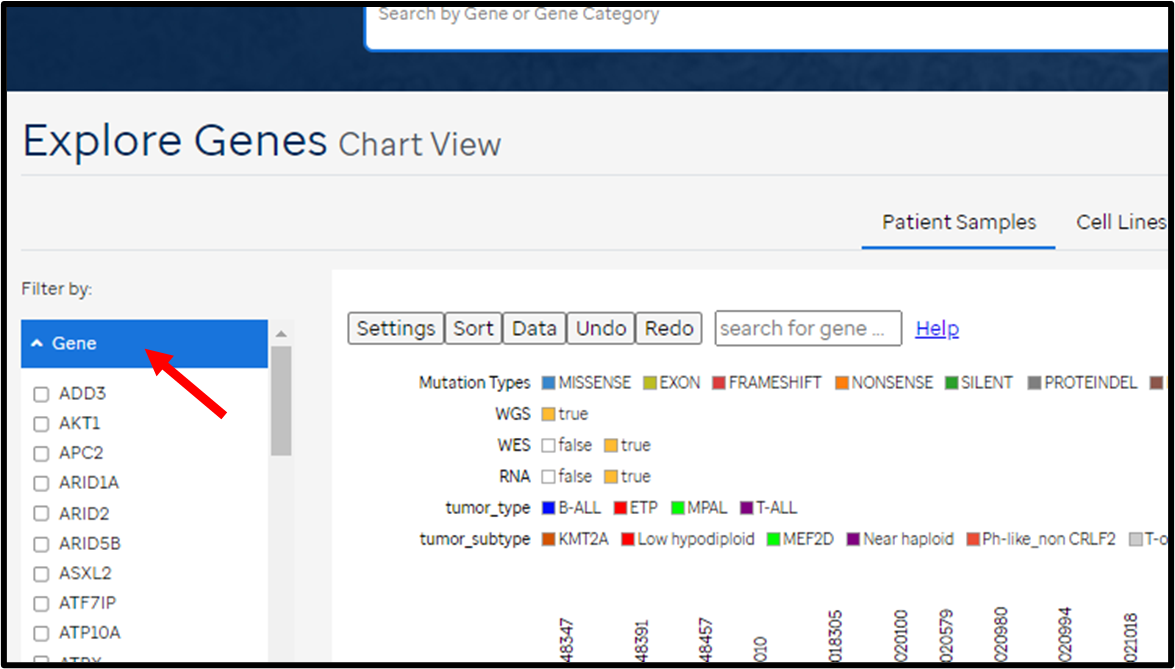
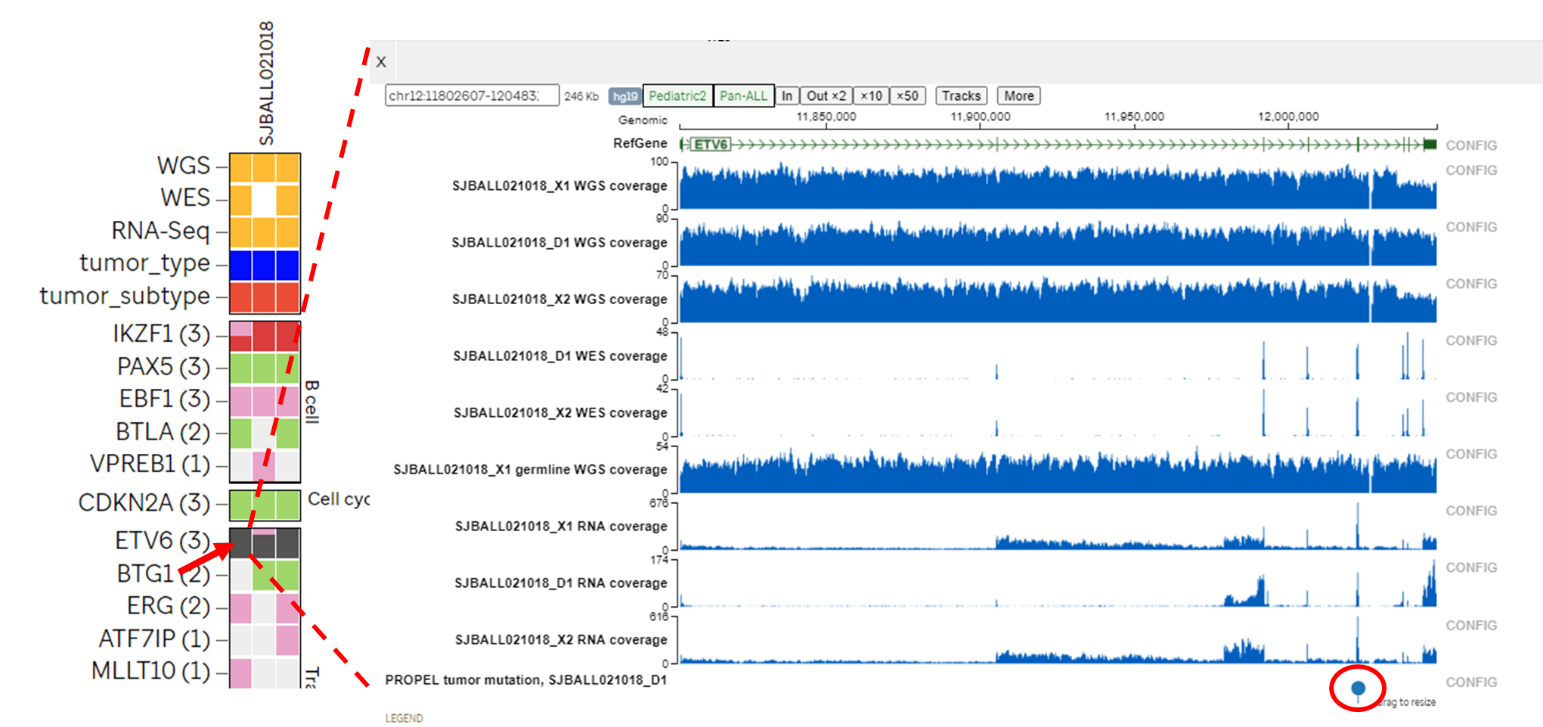
- Clicking a cell on the heatmap will bring up the Genomic Coverage page for the patient at the specific gene locus.
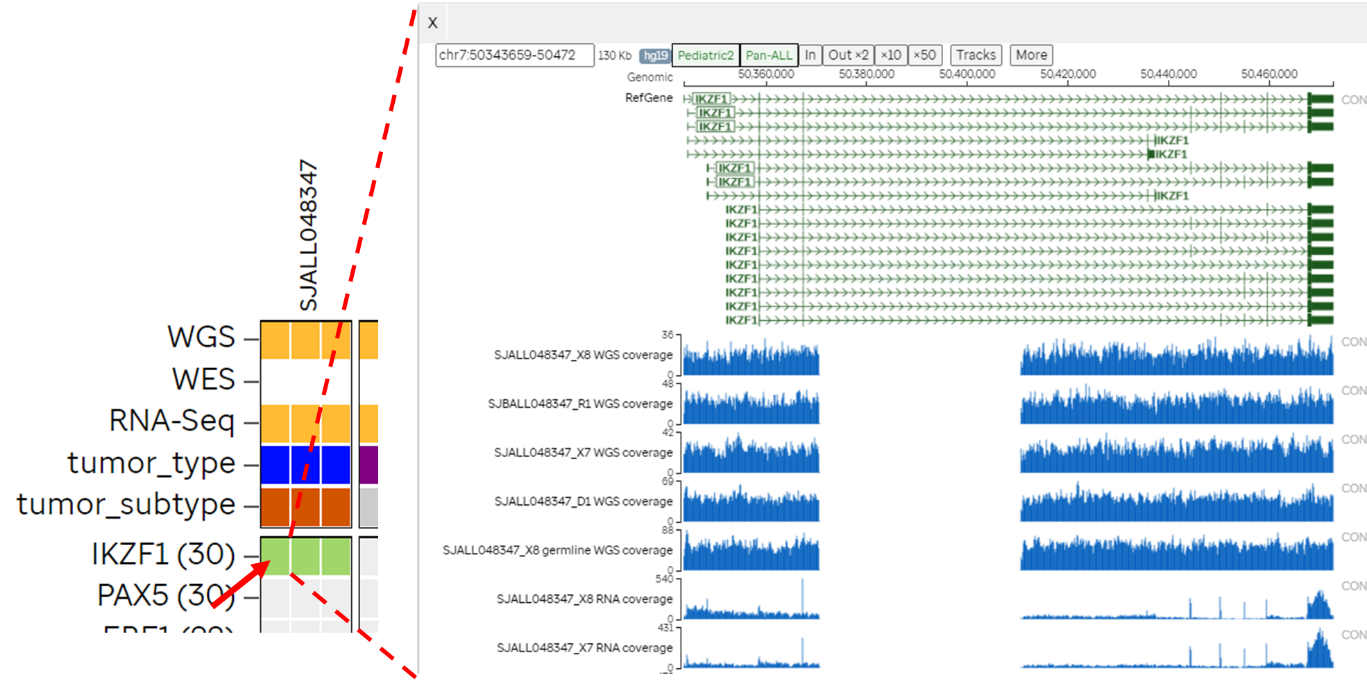
- Clicking the gene name will bring up the summary page of genomic data for the samples in this release
- Note that the cutoff for CNV in Sample Heatmap is 0.5 (absolute value of log2(CN) > 0.5), while the cutoff for CNV in genomepaint page is 0.2, which can be changed using “Config” for “Propel mutation data”.
- For aneuploid samples, and samples with whole chromosome gain/loss, CNV data only shows leukemia genes in the heatmap. Full data is displayed in genomepaint view.
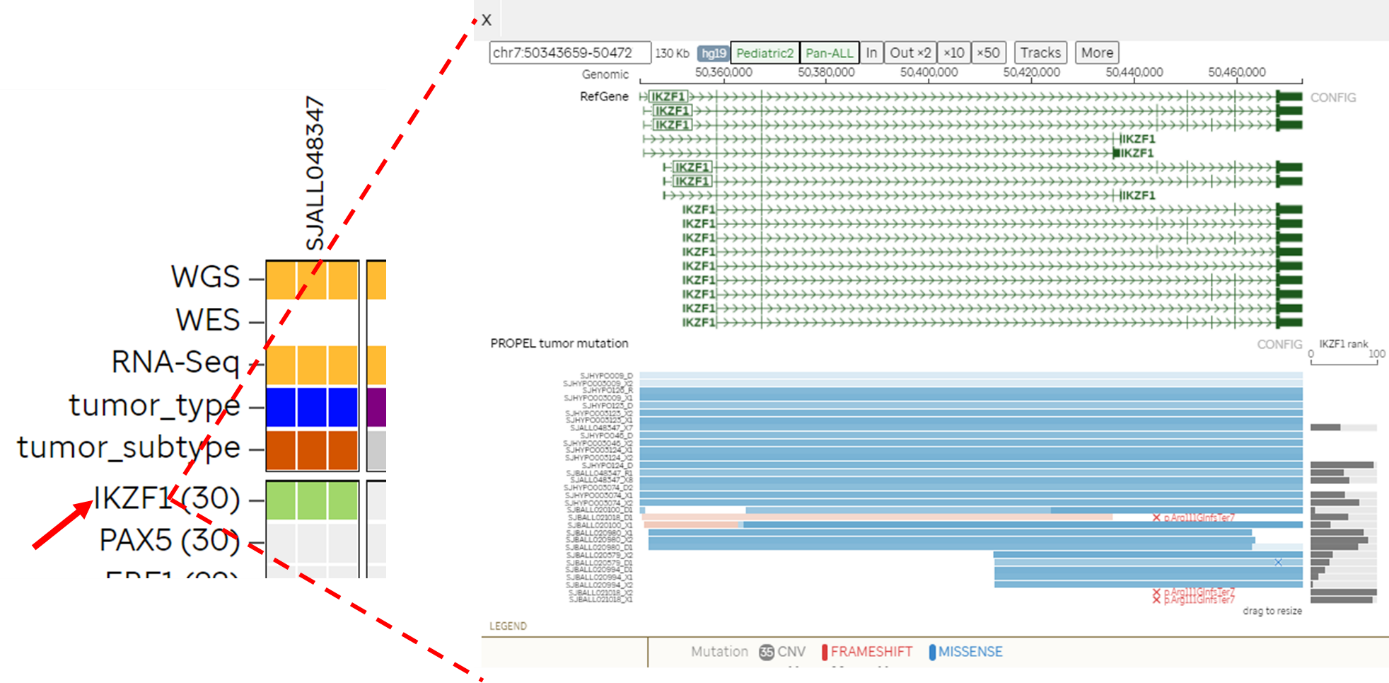
- Navigate to Sample grid under Explore Data and search for the sample of interest
- Click on the sample ID to reveal sample details
- On the left panel click on Sample Heatmap
- Choose the cell (black) that represents fusion
- In the genomepaint view, click on the blue balloon at the bottom right to see more information on the fusion partners
- Navigate to TSNE landing page under Explore Data
- Search for samples in the search bar
- Samples of a certain subtype can be highlighted by choosing the subtype listed under the search box
- More filter options can be explored from the menu in top left corner. Samples can be filtered for disease type, subtype, age, and gene rearrangementloon at the bottom right to see more information on the fusion partners
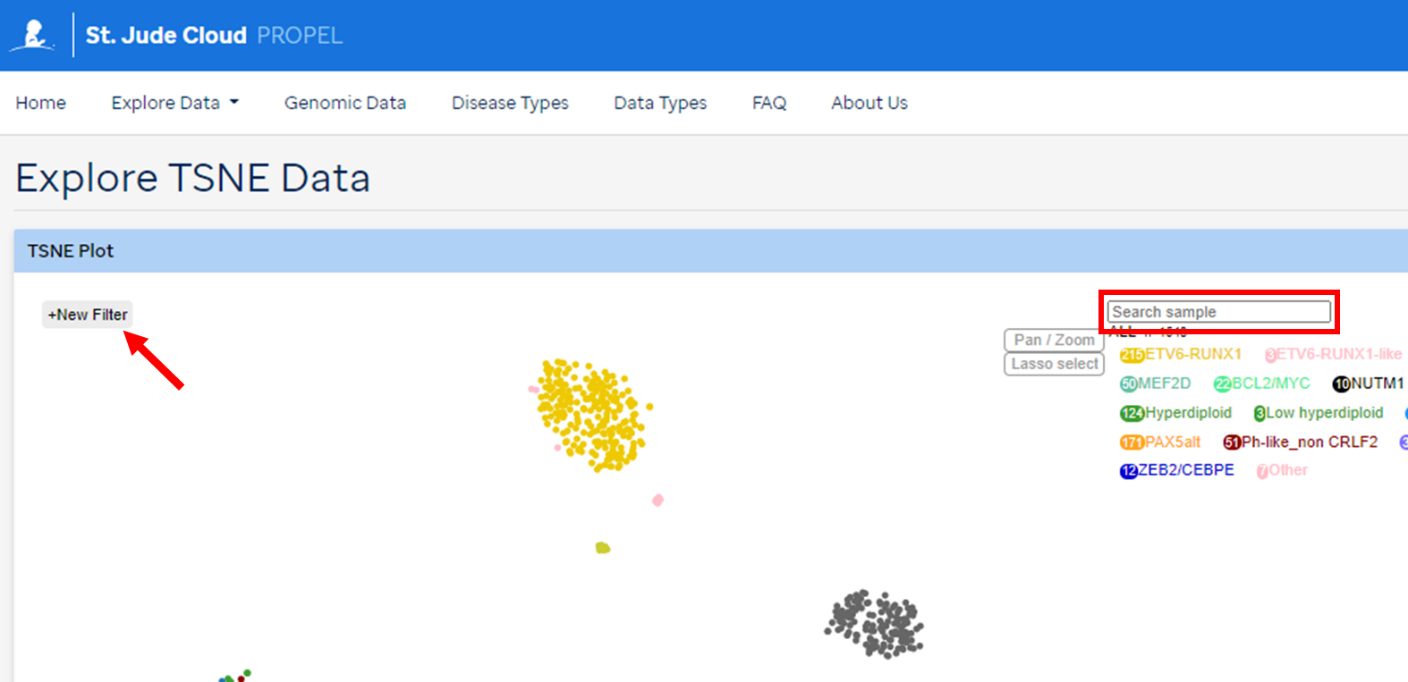
The filters allow multiple genes to be explored at the same time and the option AND/OR provides the list of samples that harbor alterations in all of them (AND) or in either of them (OR).